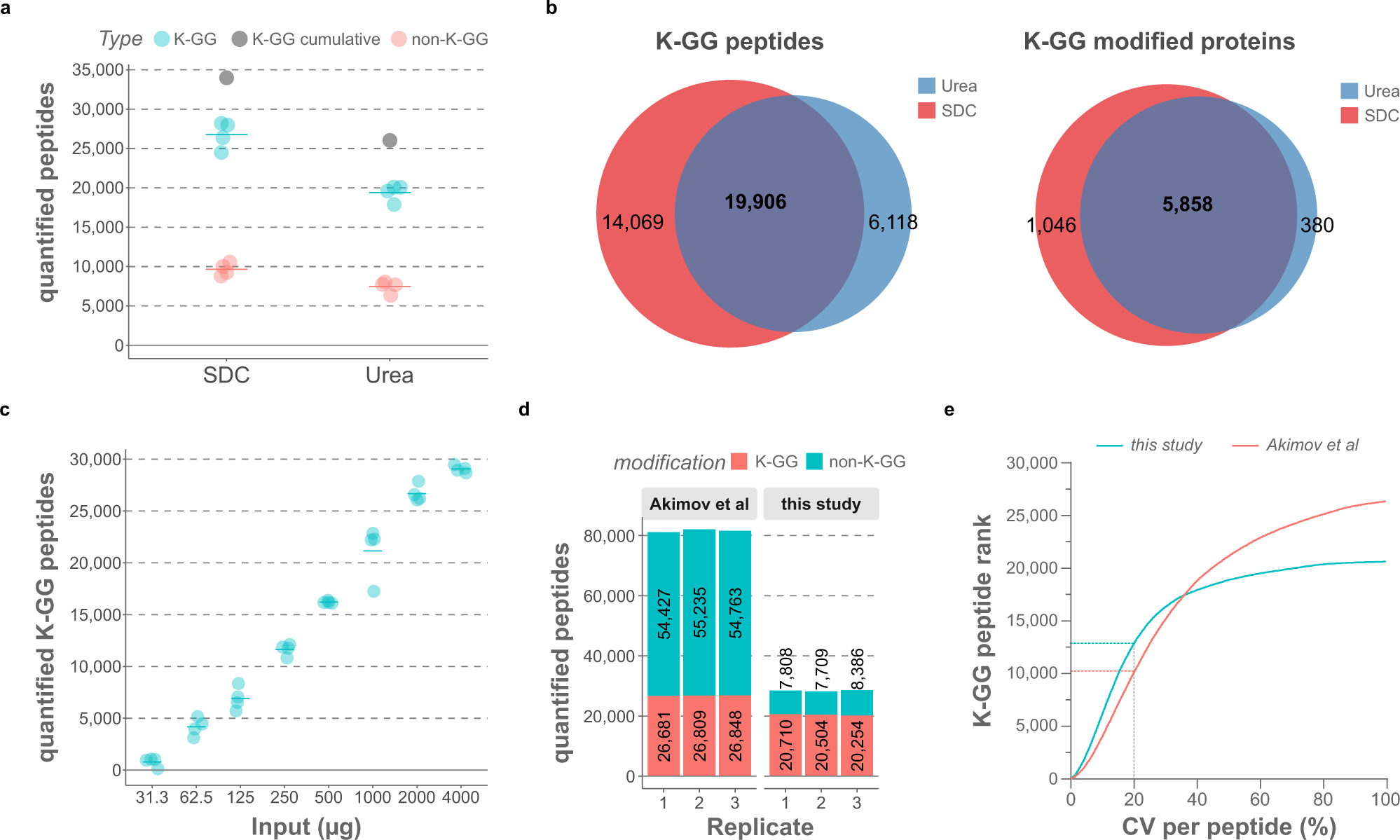
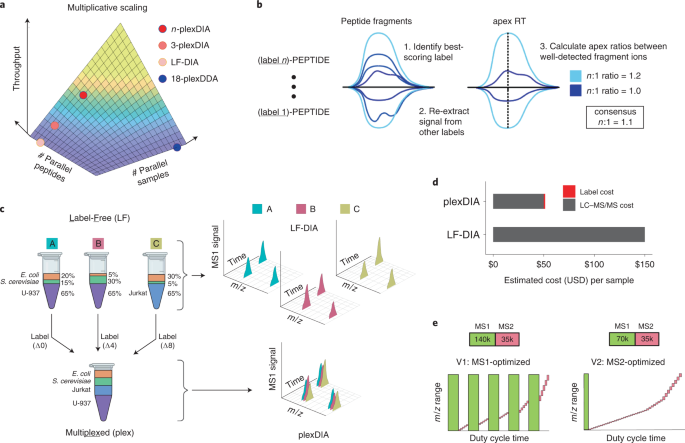
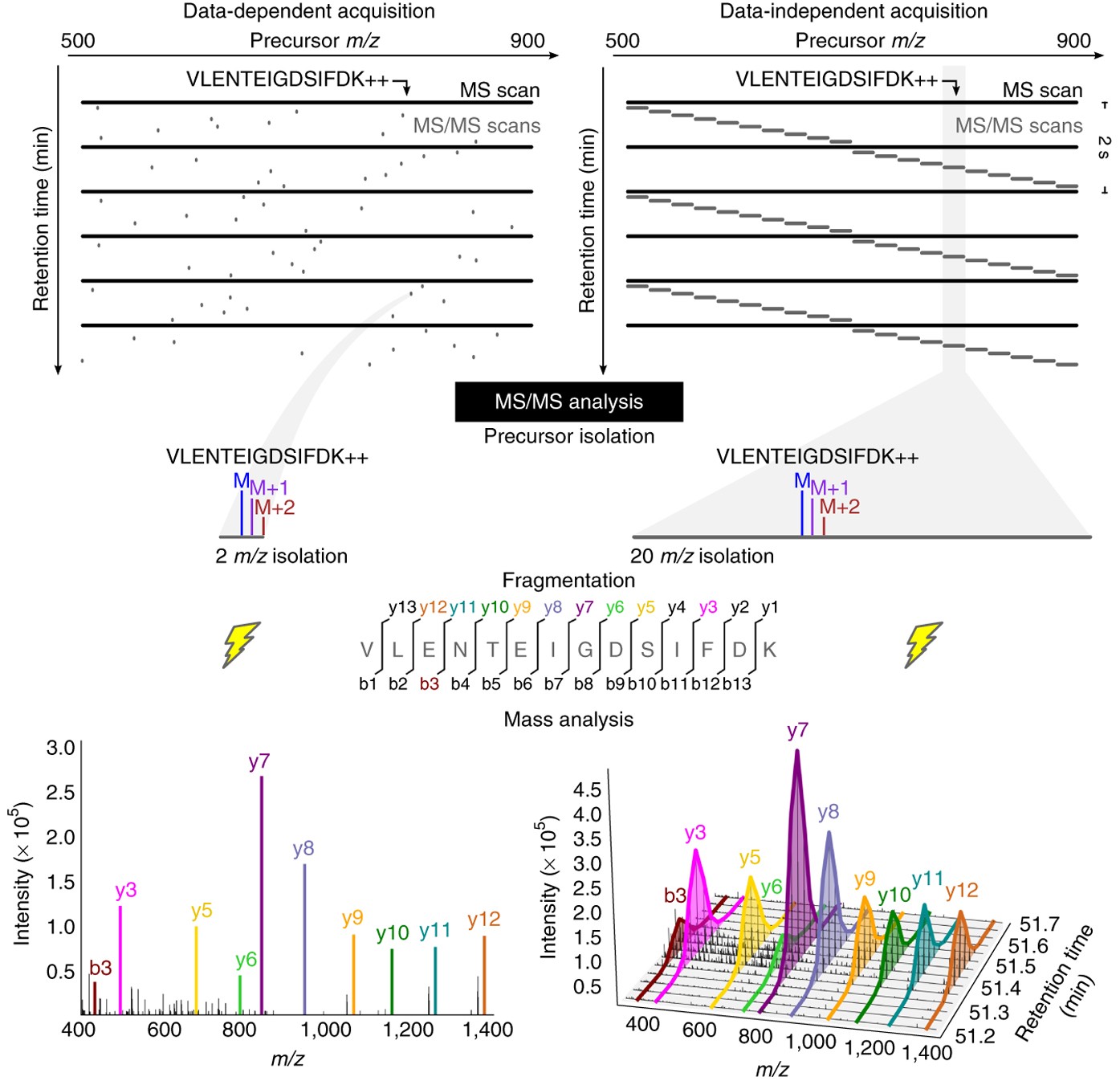
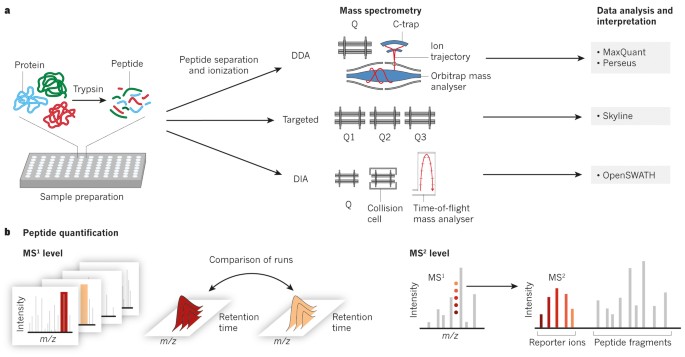
Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology | Nature Communications

Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files | Nature Methods
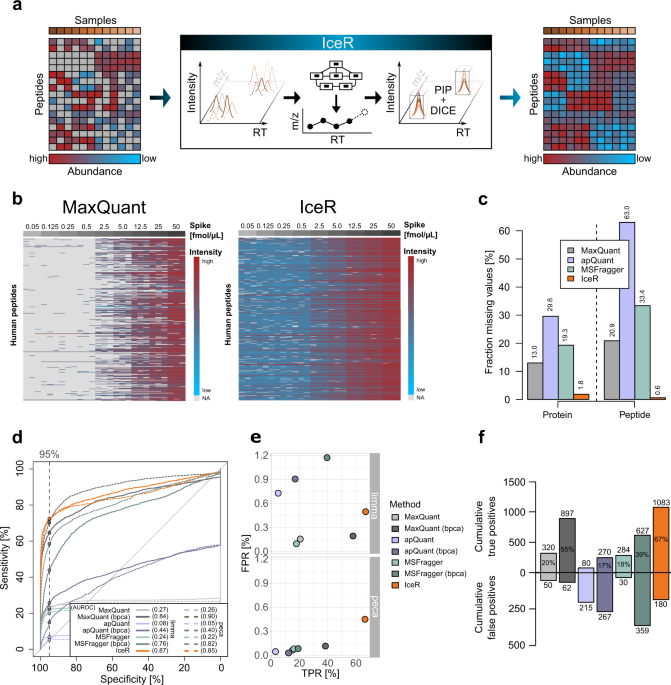
IceR improves proteome coverage and data completeness in global and single-cell proteomics | Nature Communications
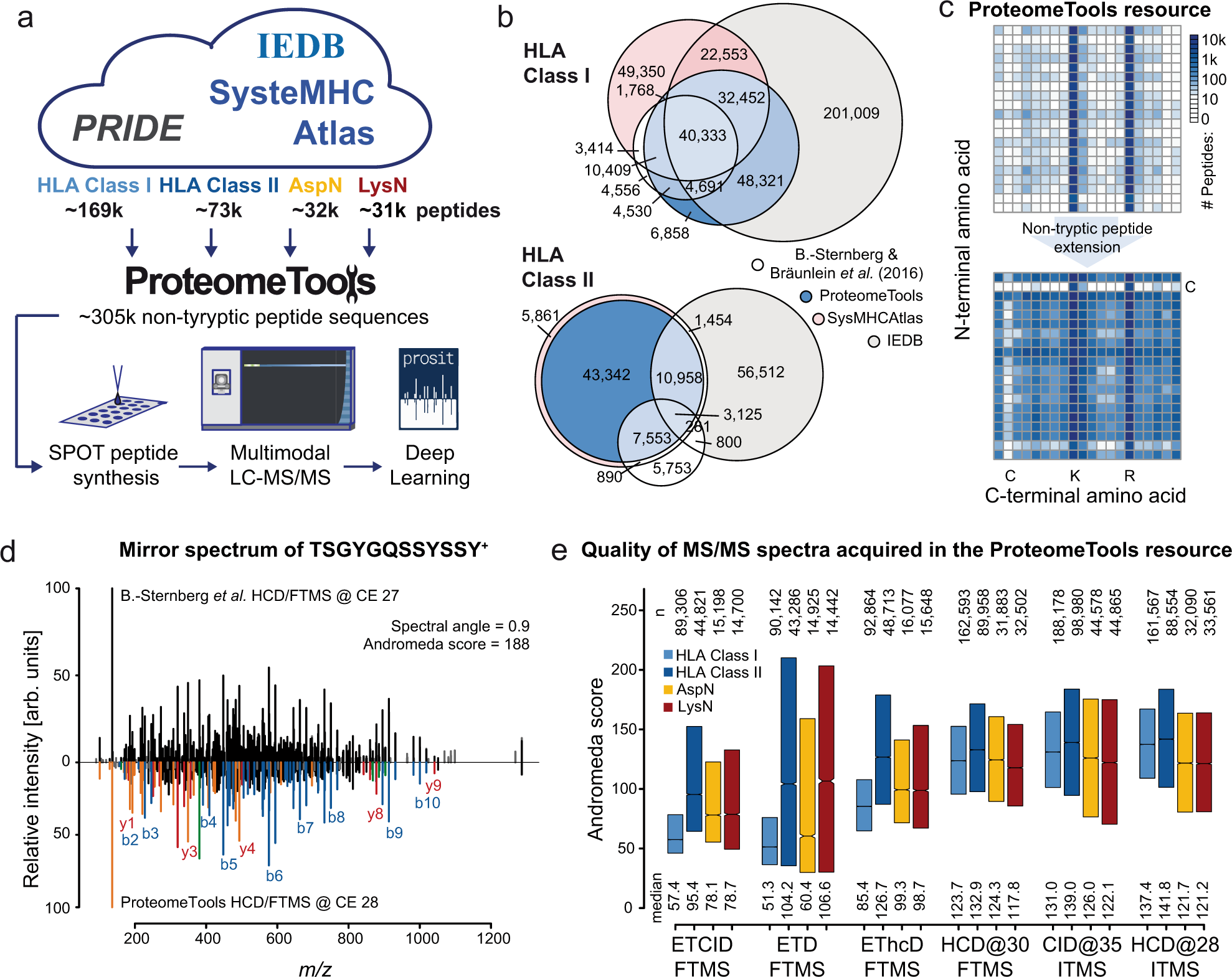
Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics | Nature Communications
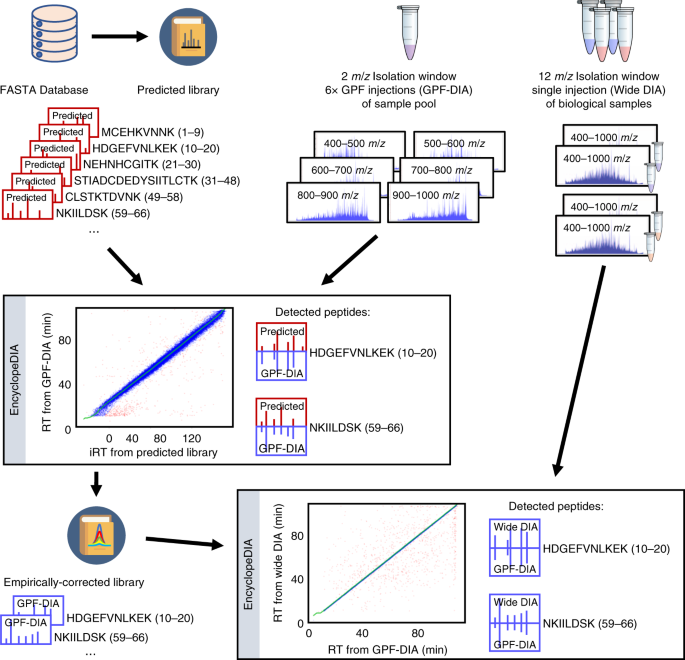
Generating high quality libraries for DIA MS with empirically corrected peptide predictions | Nature Communications
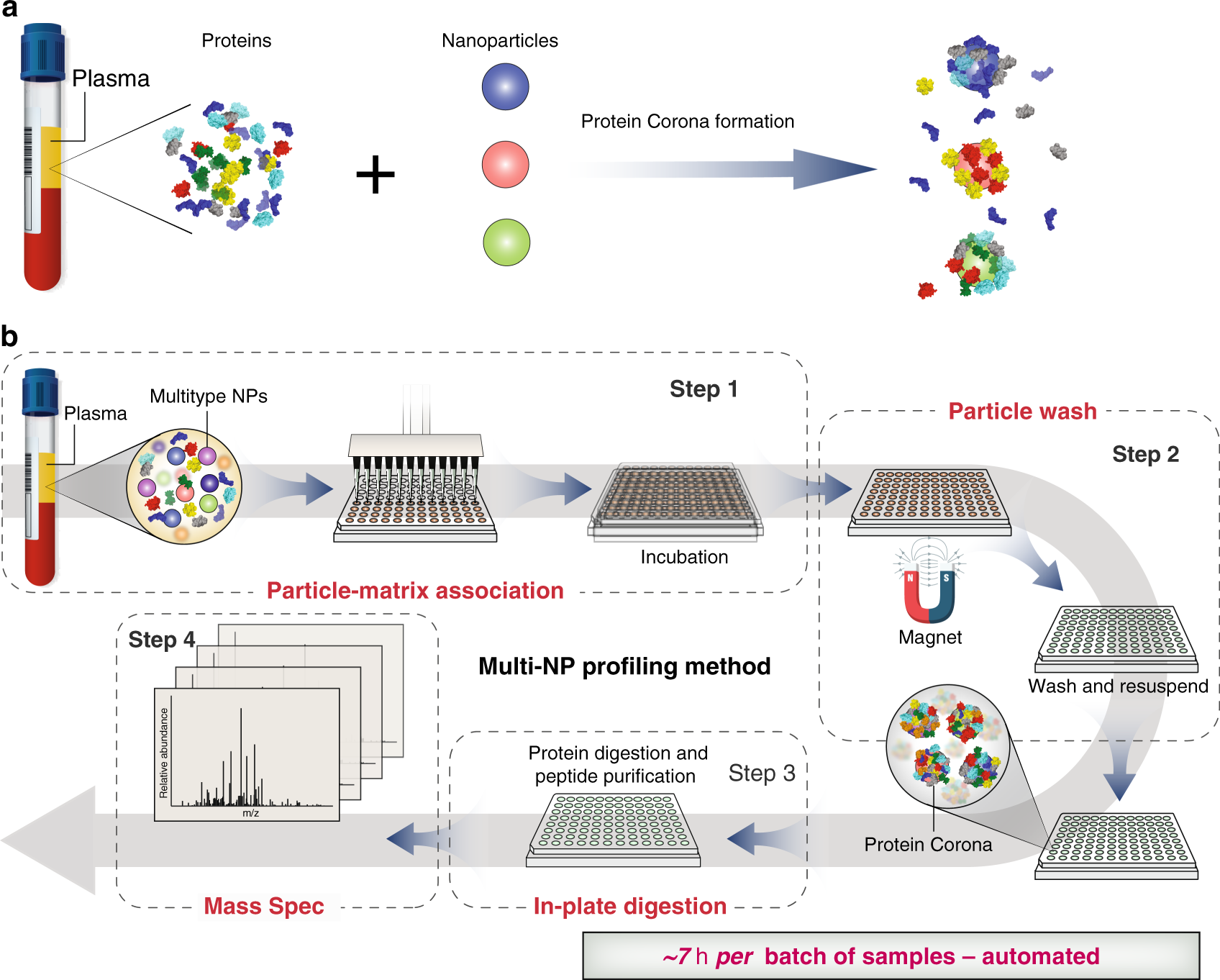
Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona | Nature Communications
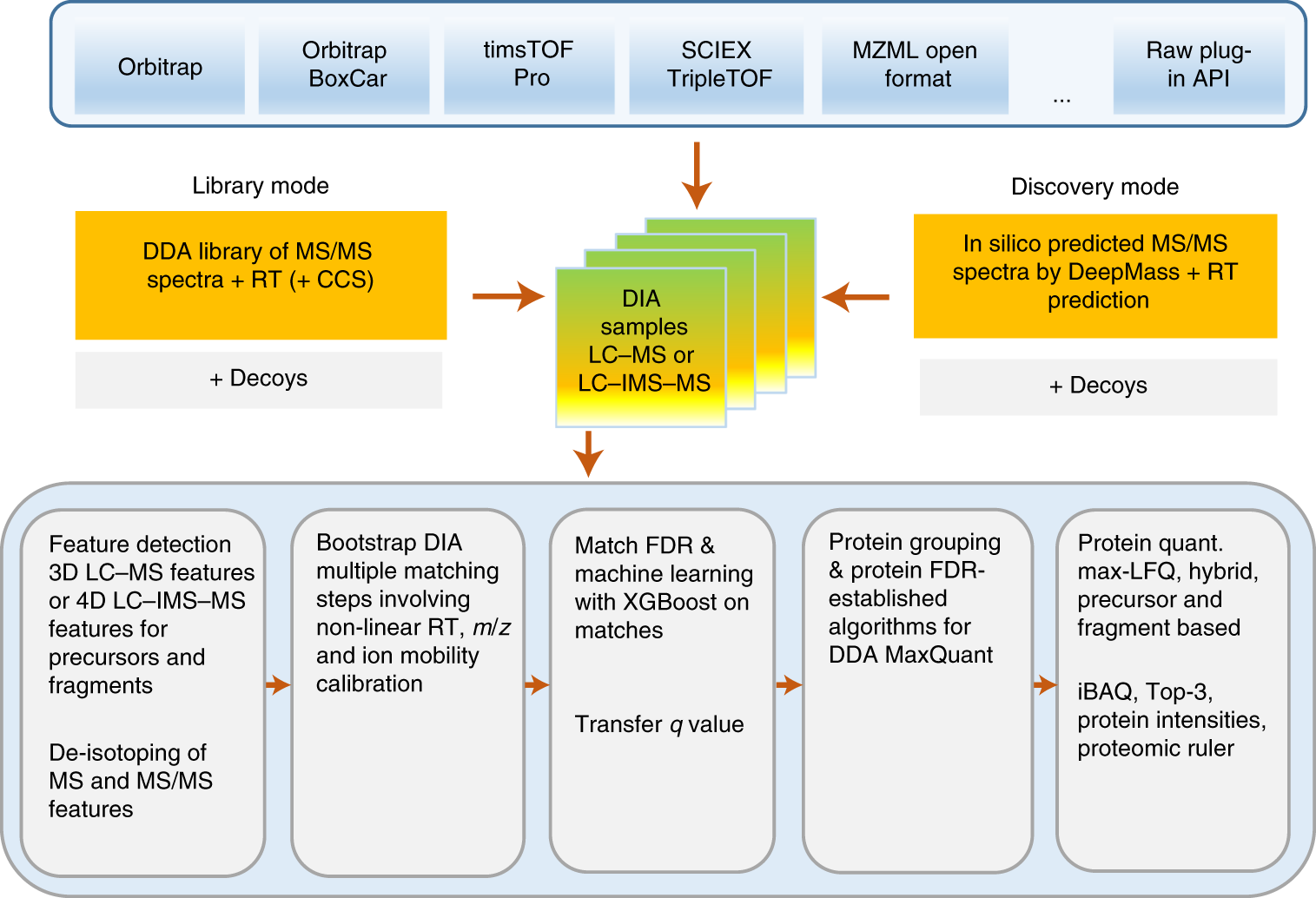
MaxDIA enables library-based and library-free data-independent acquisition proteomics | Nature Biotechnology

Deep Proteomics Using Two Dimensional Data Independent Acquisition Mass Spectrometry | Analytical Chemistry
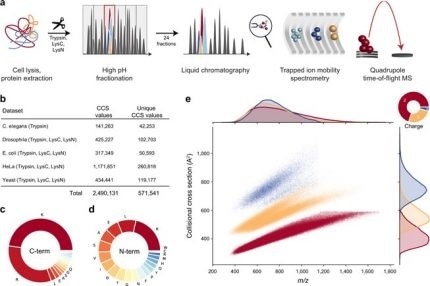
New Nature Communications publication by Mann & Theis Groups harnesses the benefits of large-scale peptide
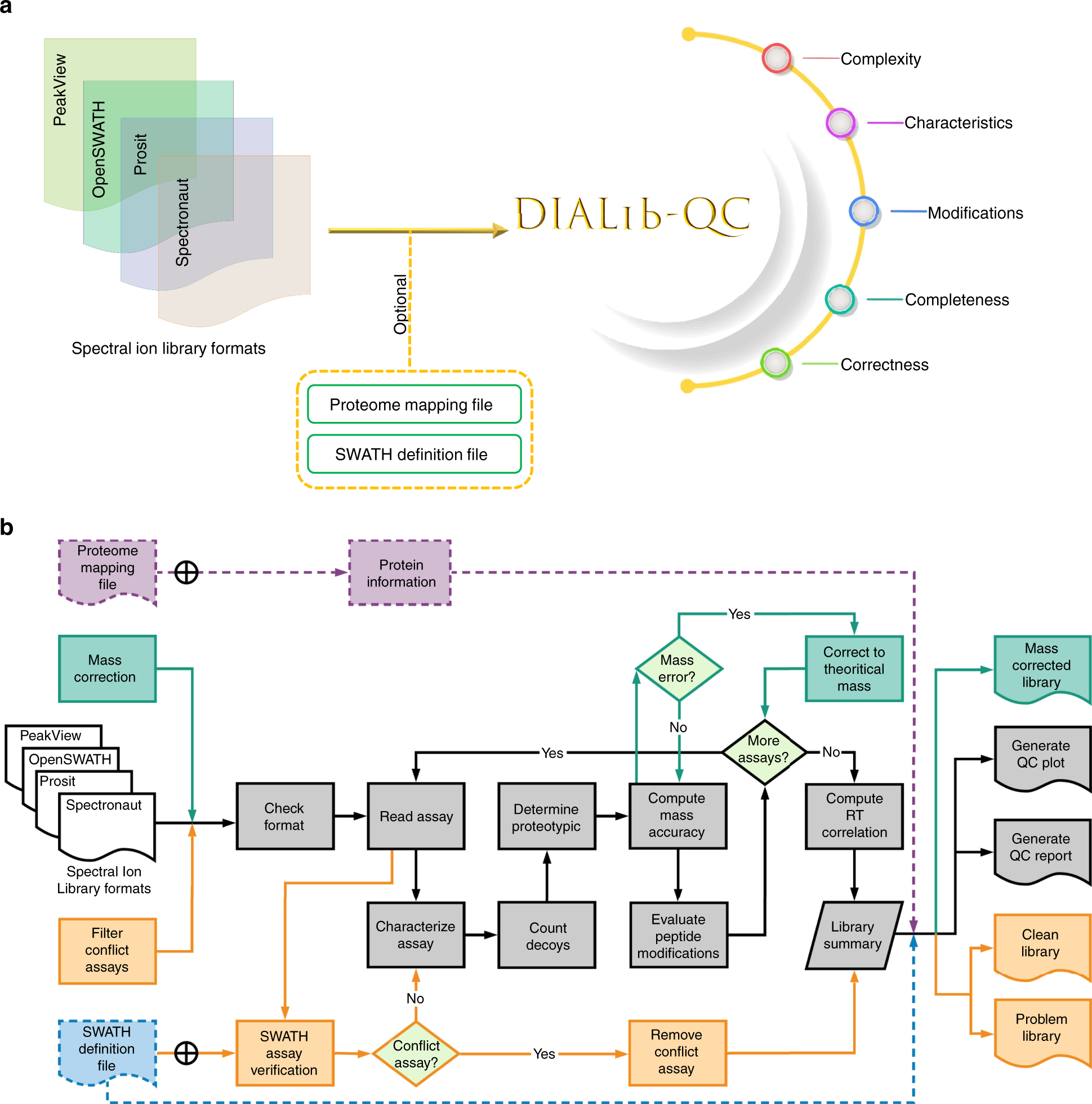
DIALib-QC an assessment tool for spectral libraries in data-independent acquisition proteomics | Nature Communications

DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput | Nature Methods
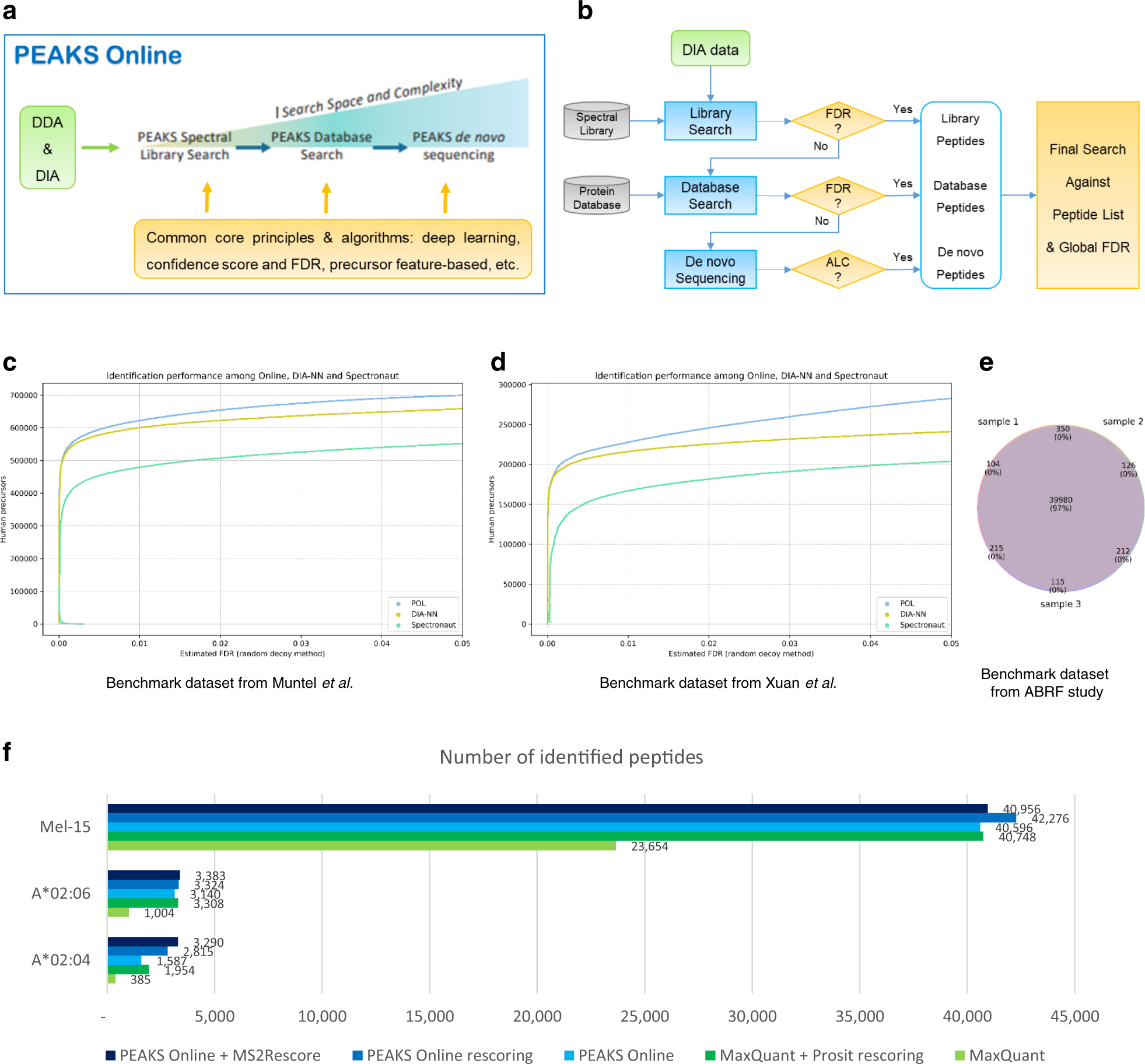
A streamlined platform for analyzing tera-scale DDA and DIA mass spectrometry data enables highly sensitive immunopeptidomics | Nature Communications

Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications
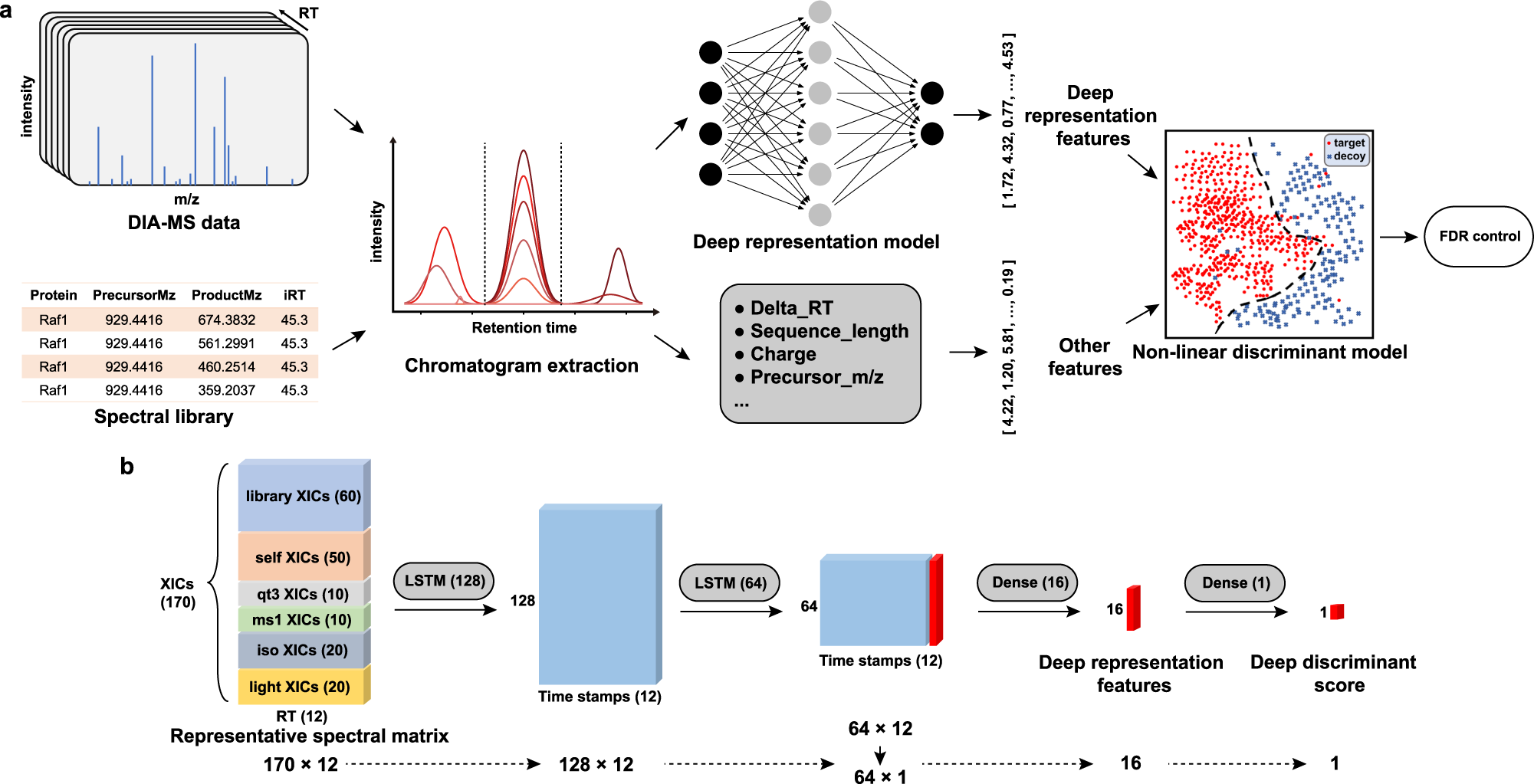
Deep representation features from DreamDIAXMBD improve the analysis of data-independent acquisition proteomics | Communications Biology
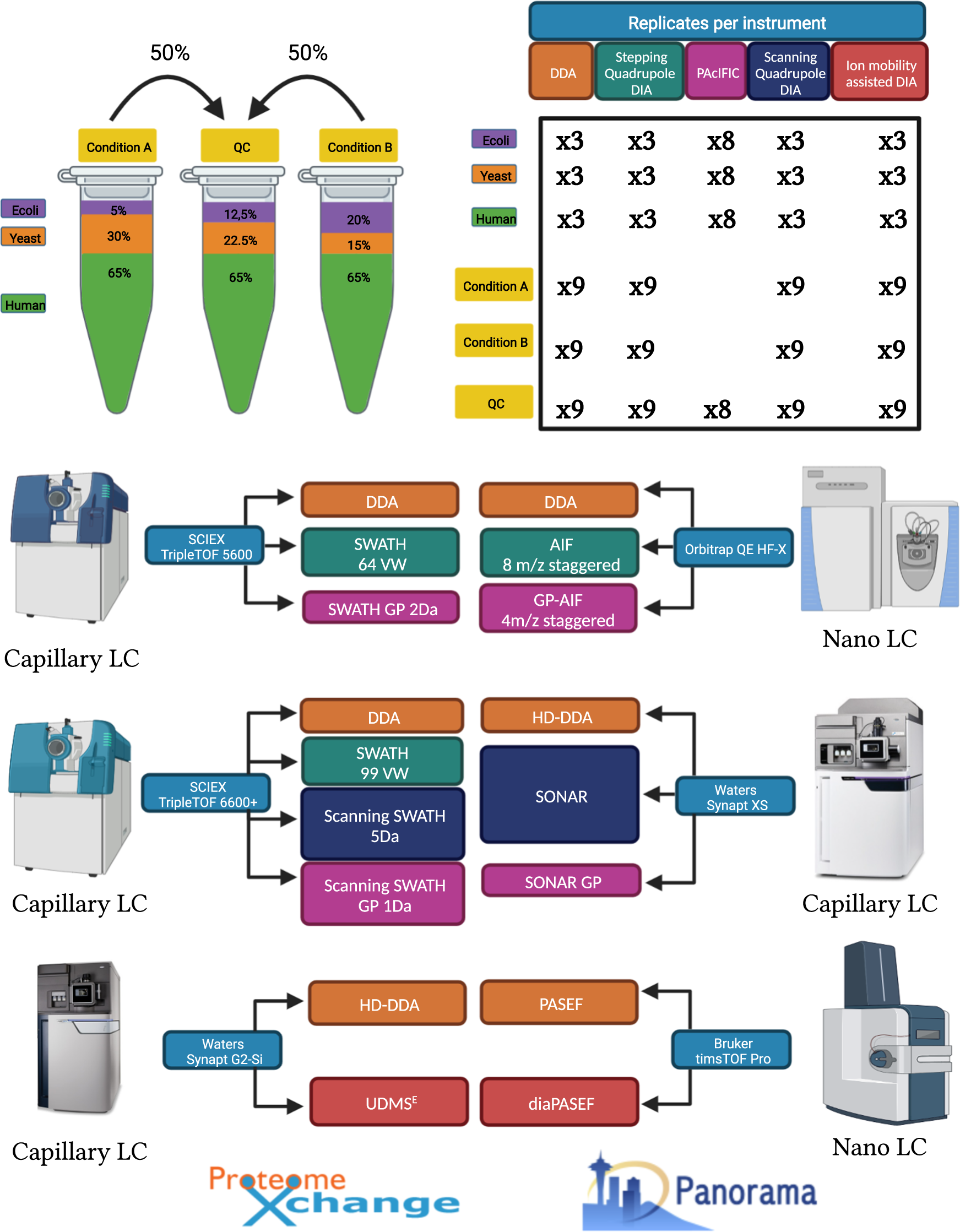
A comprehensive LFQ benchmark dataset on modern day acquisition strategies in proteomics | Scientific Data
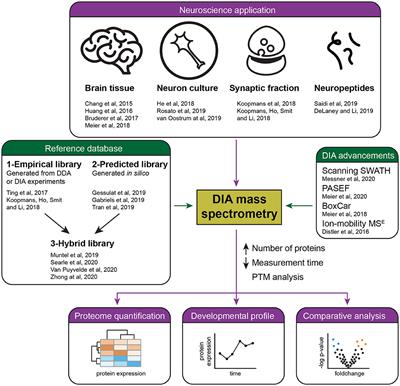
Frontiers | Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome